For a bimolecular reaction
involving two molecules A and B, with concentrations [A] and [B] and a rate
constant K, the probability of the reaction occuring (from mass action) will
be K[A][B] . Assuming a time step which is short enough to make
the probability of reaction small during the step, the mass action probability
is equal to the probability ([A]>randA && [B]>randB && K>randK). Where && is
the logical and operation, and randA, randB and randK are
uniform-distribution random numbers (Salwinski and Eisenberg and Keane, et.al.).
We can therefore replace the expensive multiplies with inexpensive logical
operations and random number generation. Random numbers were generated using
a linear
feedback shift register technique. The shift register length was chosen
so that only one xor operation was needed.
The first reaction scheme I implemented is a slightly expanded version of the system presented in Salwinski and Eisenberg. Expansions were the ability of several update events to occur for a given molecular species (as in Keane, et.al.). Examples are below. Each chemical concentration is represented by a 16-bit integer. Each reaction path can increment (+1), decrement (-1), or not change the integer concentration of a chemical at each time step. The restriction of only simple increment/decreement means that reaction probabilities must be adjusted so that the likelyhood of changing the chemical concentration by two or more is negligible. In practice the probability of reaction is set to less than 0.01, so that the probability of two events occuring is less than 0.0001. Salwinski and Eisenberg limited the update so that only one reaction could update any chemical at any time step. I added a queue so that several reactions can update a chemical at each time step. The advantage of my scheme is simpler reaction control, the disadvantage is a longer state machine on each time step. Each time step is seven clock cycles, allowing six inc/dec inputs per chemical per time step.
 More
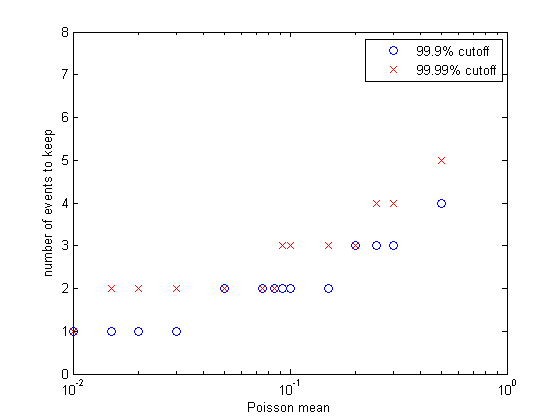
generally, what we are doing here is approximating a truncated Poisson
distribution of reaction events. We can make a better approximation by
allowing concentrations to change by more than +1/-1. Let's say that we
make the criterion that our approximation of the Poisson distribution
has to cover 99.99% of the full distribution. Or, put differently, the
cumulative sum of the discrete distribution has to be 0.9999 up to the
point we truncate. I wrote a matlab program
to plot the event number at which the cumulative probability reached
0.999 and 0.9999. The figure (left) shows the results. Keeping only one
event per reaction time step limits us to a mean rate of 0.01 at 99.99%
capture and about 0.03 at 99.9% event capture. For a mean reaction rate
up to about 0.085/time step, keeping up to two events per reaction
captures 99.99% of the events. Keeping three events allows reaction
rates up to about 0.2/time step (at 99.99% capture). So using the 99.99%
criterion, keeping two events instead of one event gives us about an
8-fold speed up, while keeping three events only gives us another factor
of two above the two-event rate.
More
generally, what we are doing here is approximating a truncated Poisson
distribution of reaction events. We can make a better approximation by
allowing concentrations to change by more than +1/-1. Let's say that we
make the criterion that our approximation of the Poisson distribution
has to cover 99.99% of the full distribution. Or, put differently, the
cumulative sum of the discrete distribution has to be 0.9999 up to the
point we truncate. I wrote a matlab program
to plot the event number at which the cumulative probability reached
0.999 and 0.9999. The figure (left) shows the results. Keeping only one
event per reaction time step limits us to a mean rate of 0.01 at 99.99%
capture and about 0.03 at 99.9% event capture. For a mean reaction rate
up to about 0.085/time step, keeping up to two events per reaction
captures 99.99% of the events. Keeping three events allows reaction
rates up to about 0.2/time step (at 99.99% capture). So using the 99.99%
criterion, keeping two events instead of one event gives us about an
8-fold speed up, while keeping three events only gives us another factor
of two above the two-event rate.
So what do we need to do to implement a maximum of two events? If we model the two reactions as separated in space, and therefore independent, we need to compute the probability of two completely independent reactions separately, then decide if 0, 1 or 2 events occured. If either reaction occurs set the inc/dec outputs to +1/-1. If both occur set the inc/dec outputs to +2/-2. If neither occurs, inc/dec are zero. This seems to be a workable system and was implemented below. A manuscript version of this description is here.
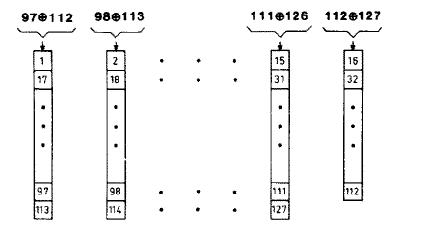
HIgh quality random number s essential for this scheme to work. It is possible to parallelize the linear feedback shift register to get more effective
shits per clock cycle. Hoogland, et.al. describe a high quality random
number generator designed for Ising model simulation. Using this 127-bit
shift register with parallel 16-bit output triples the size of the
reaction module. The following figure is taken from the paper and shows
the parallelized feedback paths of the shift register. The inputs to to
each 8-bit subsection are the xor of the two bits noted at the top of
the column. I have not yet carried out statistical tests to see if the
higher quality random numbers matter for the simulation. The 16-bit
output is taken as bits 97 to 112 from fifteen 8-bit (and one 7-bit)
shift registers. The improved Michaelis–Menten top-level module is here.
 The 127-bit generator is overkill for a 16-bit random number (but works
well for 32-bit computed in 2 cycles), so I scaled down the generator
to a 63-bit version with feedback from bits 62 and 63 (one's based). The
parallelized bit layout is the same as in the figure to the left, but
truncated at 4 bits per register.The parallelized version has sixteen
4-bit shift registers loaded in parallel. For instance, on the same
clock cycle,
The 127-bit generator is overkill for a 16-bit random number (but works
well for 32-bit computed in 2 cycles), so I scaled down the generator
to a 63-bit version with feedback from bits 62 and 63 (one's based). The
parallelized bit layout is the same as in the figure to the left, but
truncated at 4 bits per register.The parallelized version has sixteen
4-bit shift registers loaded in parallel. For instance, on the same
clock cycle,
bit62 xor bit63 is shifted into shift register 16,
bit61 xor bit62 is shifted into shift register 15,
bit48 xor bit49 is shifted into shift register 2, and
bit47 xor bit48 is shifted into shift register 1.
The Michaelis–Menten top-level module with 63-bit shift register is here. The Oregonator is here. The Oregonator example uses about 9% of the Altera DE2 board FPGA logic element resources.
The design was extended to include serial output of a waveform so
that a better comparision could be made with the Matlab differential
equation versions. The serial module takes a 16 bit number and converts
it to 4-digit hexadecimal terminated with <crlf> characters so that a call to Matlab fscan(s, '%x') can read it, where s is a serial object. The Michaelis–Menten top-level module with 127-bit shift register and serial is here (project archive). The Matlab code to read the hex is here as is the ODE analysis program (and function).
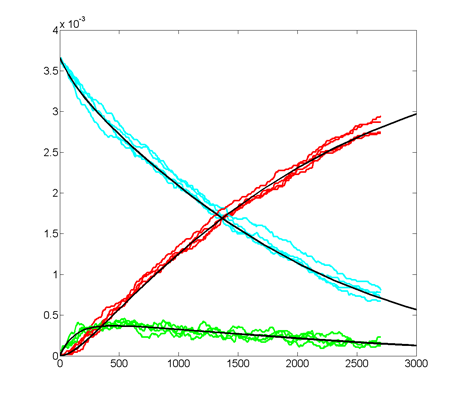
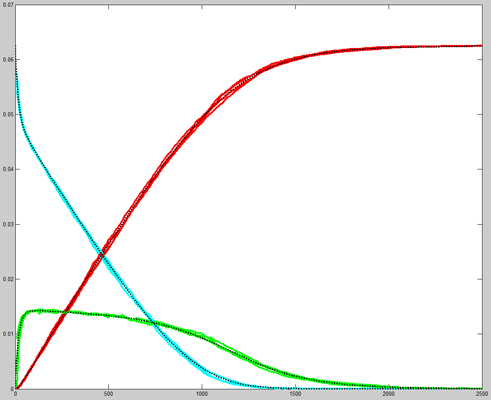
Results for substrate concentrations of 240/2^16 and 4096/2^16
are shown below. Notice the fluctuations are greater for the smaller
number of molecules on the left. The linked Verilog and Matlab programs
are coded for the higher concentration. Black lines are the Matlab
differential equation code, color lines are the FPGA output.
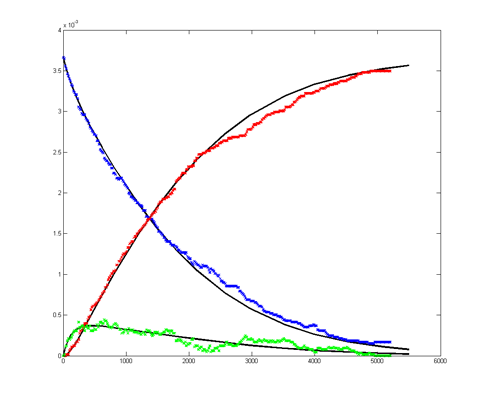
Repeating the stochastic simulations with different random number
seeds yields the following results for the two cases shown above.
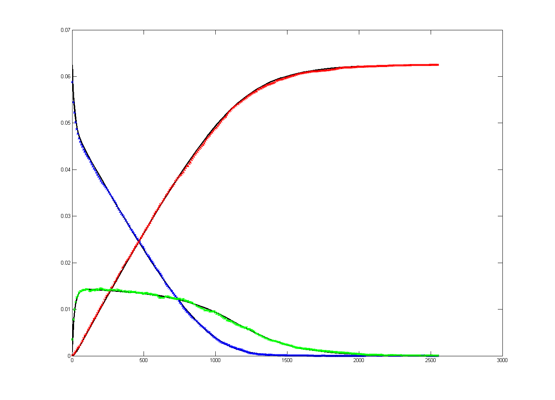
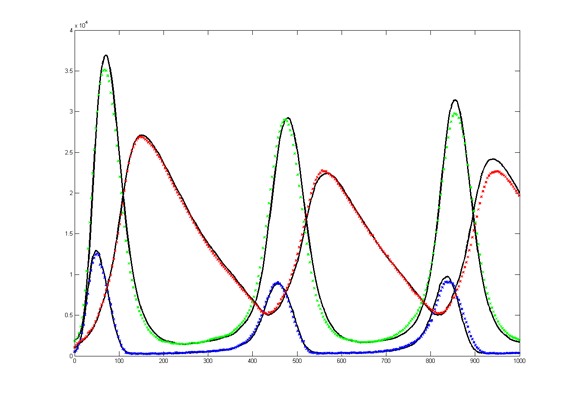
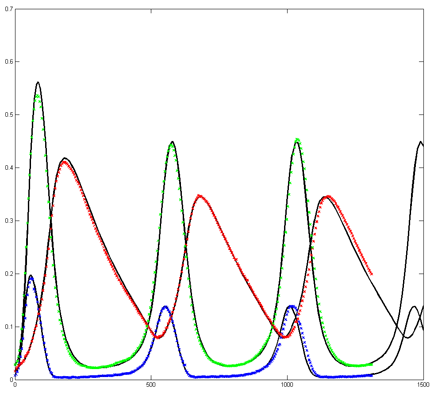
The Oregonator with 63-bit random number generator was also modified for serial output (archive). The output was compared to a Matlab simulation
of the same stochastic algorithm. The matlab version took 870 seconds
to run on my desk machine (3.2 GHz Core Duo with 8 Gbyte memory) and 8
seconds to run on the FPGA. Note that there are some significant
differences between the two simulation outputs. These diffferences seem
to depend on the random number seed used and so are probably related to
random fluctuations in chemical concentration. Black lines are the
Matlab stochastic code, color lines are the FPGA stochastic output. The
image to the right is the FPGA stochastic simulation compared to an ODE code (and function) in Matlab. In this case the amplitudes seem correct, but the period of the oscillation seems to drift.
References.
John F. Keane, Christopher Bradley, Carl Ebeling, A Compiled Accelerator for Biological Cell Signaling Simulations Cell Systems, International Symposium on Field Programmable Gate Arrays archive Proceedings of the 2004 ACM/SIGDA 12th international symposium on Field programmable gate arrays table of contents Monterey, California, USA SESSION: Pages: 233 - 241, 2004
Salwinski L, Eisenberg D., In silico simulation of biological network dynamics. Nat Biotechnol. 2004 Aug;22(8):1017-9. Epub 2004 Jul 4.
Larry Lok, The need for speed in stochastic simulation , Nature Biotechnology 22, 964 - 965 (2004) doi:10.1038/nbt0804-964
Lok L, Brent R., Automatic generation of cellular reaction networks with Moleculizer 1.0. , Nat Biotechnol. 2005 Jan;23(1):131-6.
D. Gillespie, Exact Stochastic Simulation of Coupled Chemical Reactions, Journal of Physical Chemistry, No. 81, pp. 2340-2361, 1977.
Daniel T. Gillespie, Stochastic Simulation of Chemical Kinetics, Annu. Rev. Phys. Chem. 2007.58:35-55
Hong Li,Yang Cao,Linda R. Petzold, and Daniel T. Gillespie, Algorithms and Software for Stochastic Simulation of Biochemical Reacting Systems, Biotechnol Prog. 2008; 24(1): 56–61. Published online 2007 September 26. doi: 10.1021/bp070255h.
Jürgen Pahle, Biochemical simulations: stochastic, approximate stochastic and hybrid approaches, Brief Bioinform. 2009 January; 10(1): 53–64. Published online 2009 January 16. doi: 10.1093/bib/bbn050.
R. J. Field, R. M. Noyes, Oscillations in Chemical Systems IV. Limit cycle behavior in a model of a real chemical reaction, J. Chem. Phys. 60(1974)1877-84.
A. Hoogland, J. Spaa, B. Selman and A. Compagner, A special-purpose processor for the Monte Carlo simulation of ising spin systems, Journal of Computational Physics, Volume 51, Issue 2, August 1983, Pages 250-260
 Bruce Land
Bruce Land